Workshop on Statistical Methods for Dynamic
Vancouver, June 4-6 2009
System Models
Gause's (193X) experiments on competing laboratory populations of competing species of of protozooans have been icons in ecology textbooks for generations. One experiment in particular pitted Paramecium aurelia against P. caudatum in three replicate laboratory cultures. Although both species were present in all three cultures upon the experiment's termination, Gause's makeshift parameter estimates for the Lotka-Volterra competition model suggested that P. aurelia was well on its way to competitive victory in all three cultures. The resulting `Principle of Competitive Exclusion' of one species by a close competitor has infused ecological and evolutionary thought ever since.
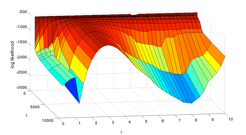
Astonishingly, Gause's original data have not received much analysis to date in the ecological literature. One problem has been that the population abundances in the data display considerable variability. Tractable methods of fitting competition models to the data (i.e. estimating model parameters) have assumed that the variability either is entirely sampling error arising from Gause's laboratory methods or is entirely natural biological variability ('process noise') in the growth rates of the populations themselves. An additional problem is that the time series data have missing values. Classical statistical inferences for realistic models accounting for combined sources of variability and missing data have not been possible, because until recently calculating the likelihood functions for such `state space' models has not been computationally feasible.
We used the new 'data cloning' algorithm (Lele et al. 2007) to conduct statistical inferences for Gause's P. aurelia v. P. caudatum experiment. The inferences are based on a full state-space competition model featuring both sampling error and process noise. The base model is the discrete time Leslie-Gower competition model which has the same dynamical behavior and stability properties (linear crossed isoclines, etc.) as the Lotka-Volterra model. Maximum likelihood parameter estimates put the populations in a dynamical region of coexistence, that is, the estimated model predicts that the populations will ultimately attain a stable positive equilibrium. The model was refitted using data cloning to 100 data sets simulated from the original model and ML estimates. Of these 100 bootstrap data sets, 67 of them predicted coexistence, and the remaining 33 predicted competitive exclusion of P. caudatum by P. aurelia. Adequate modeling of the stochastic variability as well as the deterministic forces is critical for understanding population dynamical behavior.
Authors: Brian Dennis and Subhash R. Lele
Gause's principle of competitive coexistence?
Professor of Wildlife and Statistics
Department of Fish and Wildlife Resources
University of Idaho
Moscow Idaho